PRIMER REPORTE DE DIAGNÓSTICO MOLECULAR DE DIARREA EPIDÉMICA PORCINA EN ECUADOR
Contenido principal del artículo
Resumen
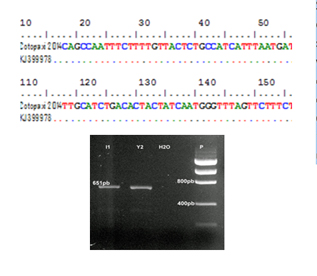
Se presentan los resultados de la confirmación por RT-PCR y secuenciación de la presencia del virus de la diarrea epidémica porcina en cerdos con diarrea por primera vez en Ecuador la secuencia del gen S de Cotopaxi 2014, tuvo una identidad de 100% con cepas o aislados en Estados unidos
Descargas
La descarga de datos todavía no está disponible.
Detalles del artículo
Cómo citar
1.
Ana Garrido, Maritza Barrera, María Vaca, Alfredo Acosta. PRIMER REPORTE DE DIAGNÓSTICO MOLECULAR DE DIARREA EPIDÉMICA PORCINA EN ECUADOR. EEC [Internet]. 29 de agosto de 2015 [citado 10 de marzo de 2026];2(2). Disponible en: https://revistaecuadorescalidad.agrocalidad.gob.ec/revistaecuadorescalidad/index.php/revista/article/view/10
Sección
Artículos Científicos
Citas
E.N. Wood (1977) “An apparently new syndrome of porcine epidemic diarrhea”,Vet Rec.100, 243–4.
R.J. De Groot, S.C. Baker, R. Baric, L. Enjuanes, A.E. Gorbalenya, K.V. Holmes, S. Perlman, L. Poon, P.J.M. Rottier,P.J. Talbot, P.C.Y. Woo, J..Ziebuhr (2011),"Family Coronaviridae", En: “Ninth Report of the International Committee on Taxonomy of Viruses”,(Ed.:A.M.Q. King, E.Lefkowitz, M.J. Adams, E.B. Carstens), Elsevier, Oxford, pp. 806-828.
D.S. Song, J.B.K. Park (2012) “Porcine epidemic diarrhea virus: a comprehensive review ofmolecular epidemiology, diagnosis, and vaccines”. Virus Genes, 44,167–75.
L.Enjuanes, D.Cavanagh. (2002) “Coronaviridae”, En:"The Springer Index of Viruses",(Eds.:C. A
Tidona,G.Darai), University of Heidelberg, Alemania,pp, 272-280.
S. Lee, C. Lee (2014) “Outbreak-Related Porcine Epidemic Diarrhea Virus Strains Similar toUS Strains, South Korea, 2013”, Emerging Infect. Dis.20 (7), 1223-1226.
Y.W.Huang, A.W.Dickerman, P. Pineyro,L.Li, .Fang, R .Kiehne (2013) “Origin, evolution and genotyping of emergent porcine epidemic diarrhea virus strains in the United States”. mBio.4, e00737–13.
Y.Tian , Z. Yu, K..Cheng,Y. Liu , J. Huang, Y. Xin , Y. Li , S. Fan , T. Wang , G .Huang , N. Feng , Z. Yang , S. Yang, Y Gao, X.Xia (2013) “Molecular Characterization and Phylogenetic Analysis of New Variants of the Porcine Epidemic Diarrhea Virus in Gansu, China in 2012”,Viruses, 5, 1991-2004; doi:10.3390/v5081991.
A.N.Vlasova, D.Marthaler, Q. Wang, M.R Culhane., K.D.Rossow,A.Rovira, J. Collins, L.J.Saif (2014) “Distinct Characteristics and Complex Evolution of PEDV Strains,North America, May 2013-February 2014”, Emerging Infect. Dis. 20 (10), 1620-1628.
L Wang, B Byrum, Y Zhang (2014) “New Variant of Porcine Epidemic Diarrhea Virus, United States 2014. Emerging Infectious Diseases; 20 (5), 918-919.
K. Jung, Q. Wang, K.A. S. Zhongyan Lu, Y. Zhang, L.J. Saif (2014). “Pathology of US Porcine Epidemic Diarrhea Virus Strain PC21A in Gnotobiotic Pigs”. Emerging Infect. Dis. 20(4), 662-665.
D.S.Song,K.B.Kang, J.S.Oh, W.Gun, J.S.Yang,H.J. Moon, K.Yong-Suk, J.B.K.Park ( 2006) “Multiplex reverse transcription-PCR for rapid differential detection of porcine epidemic diarrhea virus, transmissible gastroenteritis virus, and porcine group A rotavirus”,J.Vet.Diagn. Invest.18, 278–281.
T.A.Hall (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41, 95–98.
K. Tamura, G. Stecher, D. Peterson, A. Filipski, S. Kumar (2013) “MEGA6: Molecular Evolutionary
Genetics Analysis version 6.0”. Molecular Biology and Evolution. 30, 2725-2729.
T. Oka,L.J. Saif,D. Marthaler,M.A. Esseilia,T. Meuliad,.C.M. Lin,A.NVlasova,K. Jung,Y. Zhang,Q.Wang ( 2014)“Cell culture isolation and sequence analysis of genetically diverse US porcine
epidemic diarrhea virus strains including a novel strain with a large deletion in the spike gene”,Vet.
Microbiol.173(3-4),258–269.
R.J. De Groot, S.C. Baker, R. Baric, L. Enjuanes, A.E. Gorbalenya, K.V. Holmes, S. Perlman, L. Poon, P.J.M. Rottier,P.J. Talbot, P.C.Y. Woo, J..Ziebuhr (2011),"Family Coronaviridae", En: “Ninth Report of the International Committee on Taxonomy of Viruses”,(Ed.:A.M.Q. King, E.Lefkowitz, M.J. Adams, E.B. Carstens), Elsevier, Oxford, pp. 806-828.
D.S. Song, J.B.K. Park (2012) “Porcine epidemic diarrhea virus: a comprehensive review ofmolecular epidemiology, diagnosis, and vaccines”. Virus Genes, 44,167–75.
L.Enjuanes, D.Cavanagh. (2002) “Coronaviridae”, En:"The Springer Index of Viruses",(Eds.:C. A
Tidona,G.Darai), University of Heidelberg, Alemania,pp, 272-280.
S. Lee, C. Lee (2014) “Outbreak-Related Porcine Epidemic Diarrhea Virus Strains Similar toUS Strains, South Korea, 2013”, Emerging Infect. Dis.20 (7), 1223-1226.
Y.W.Huang, A.W.Dickerman, P. Pineyro,L.Li, .Fang, R .Kiehne (2013) “Origin, evolution and genotyping of emergent porcine epidemic diarrhea virus strains in the United States”. mBio.4, e00737–13.
Y.Tian , Z. Yu, K..Cheng,Y. Liu , J. Huang, Y. Xin , Y. Li , S. Fan , T. Wang , G .Huang , N. Feng , Z. Yang , S. Yang, Y Gao, X.Xia (2013) “Molecular Characterization and Phylogenetic Analysis of New Variants of the Porcine Epidemic Diarrhea Virus in Gansu, China in 2012”,Viruses, 5, 1991-2004; doi:10.3390/v5081991.
A.N.Vlasova, D.Marthaler, Q. Wang, M.R Culhane., K.D.Rossow,A.Rovira, J. Collins, L.J.Saif (2014) “Distinct Characteristics and Complex Evolution of PEDV Strains,North America, May 2013-February 2014”, Emerging Infect. Dis. 20 (10), 1620-1628.
L Wang, B Byrum, Y Zhang (2014) “New Variant of Porcine Epidemic Diarrhea Virus, United States 2014. Emerging Infectious Diseases; 20 (5), 918-919.
K. Jung, Q. Wang, K.A. S. Zhongyan Lu, Y. Zhang, L.J. Saif (2014). “Pathology of US Porcine Epidemic Diarrhea Virus Strain PC21A in Gnotobiotic Pigs”. Emerging Infect. Dis. 20(4), 662-665.
D.S.Song,K.B.Kang, J.S.Oh, W.Gun, J.S.Yang,H.J. Moon, K.Yong-Suk, J.B.K.Park ( 2006) “Multiplex reverse transcription-PCR for rapid differential detection of porcine epidemic diarrhea virus, transmissible gastroenteritis virus, and porcine group A rotavirus”,J.Vet.Diagn. Invest.18, 278–281.
T.A.Hall (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41, 95–98.
K. Tamura, G. Stecher, D. Peterson, A. Filipski, S. Kumar (2013) “MEGA6: Molecular Evolutionary
Genetics Analysis version 6.0”. Molecular Biology and Evolution. 30, 2725-2729.
T. Oka,L.J. Saif,D. Marthaler,M.A. Esseilia,T. Meuliad,.C.M. Lin,A.NVlasova,K. Jung,Y. Zhang,Q.Wang ( 2014)“Cell culture isolation and sequence analysis of genetically diverse US porcine
epidemic diarrhea virus strains including a novel strain with a large deletion in the spike gene”,Vet.
Microbiol.173(3-4),258–269.